Computational Modelling
Our lab uses modelling techniques in parallel with experimental investigations; a powerful combination enabling a quantitative understanding of various complex biological systems . These modelling techniques include classical density functional theory (DFT), in collaboration with Ian Ford, and course-grained molecular dynamics (MD), in collaboration with Andela Saric, as well as Brownian dynamics and rate equation models. In addition to simulations, we use various computational algorithms to aid the analysis of our experimental data.
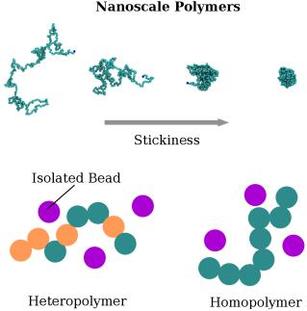
Theoretical and computational modelling allows the scientist to probe a biological system, through simulations, in ways that experimental techniques cannot. Usually the biological system is represented in a more simplified way so that principles from physics and mathematics can be used to gain insight into the system. For example a disordered protein in the nuclear pore complex could be represented as a chain of beads (polymer), with other molecules represented as isolated beads.
