Nuclear Pore Complex
|
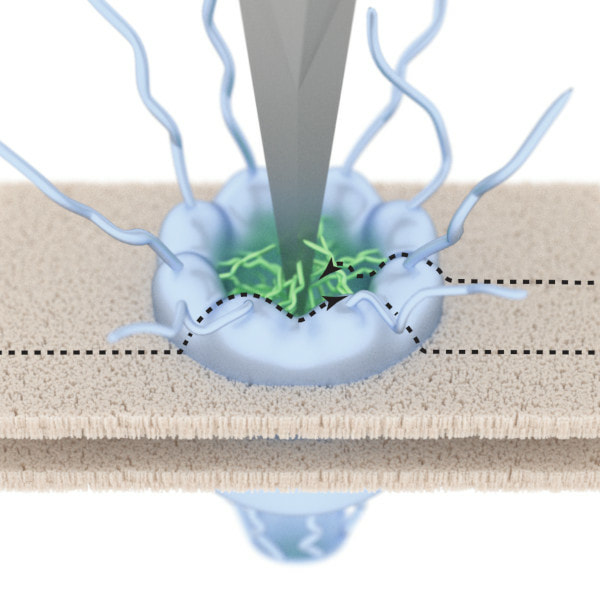
At the heart of every cell in our body is a cell nucleus, a dense structure that contains our DNA. For a cell to function normally, it needs to surround its nucleus with a protective envelope, and yet this envelope must open enough to let vital molecules in and out. To this end, it is pierced by hundreds of tiny gateways known as nuclear pore complexes (NPCs). In the NPC, an assembly of natively unfolded ("spaghetti-like") proteins determines the chemical and size selectivity of transport into and out of the nucleus. We use a variety of (bio)physical methods to understand how this protein assembly can create a size-selective transport barrier that yet can let specific large cargos (e.g., some viruses) get through. In some of our approaches, we gently poke the transport barrier with an AFM tip. In others, we develop computational models based on polymer physics for comparison with and thus interpretation of experimental data.
|
|
|
We have a strong record of using reconstituted assemblies of nuclear pore proteins as model systems that enable us to probe NPC-relevant behaviour under highly controlled conditions. This is largely thanks to collaborations with Ralf Richter in Leeds, and with Chenxiang Lin and Patrick Lusk at Yale. With the Yale team, we have for example visualised collective protein dynamics in NPC mimics based on DNA origami nanopores.
|
Related Publications
- L. K. Davis, I. J. Ford and B. W. Hoogenboom, eLife, 2022, 11, e72627
- P. D. Ellis Fisher, Q. Shen, B. Akpinar, L. K. Davis, K. K. H. Chung, D. Baddeley, A. Saric, T. J. Melia, B. W. Hoogenboom, C. Lin and C. P. Lusk, ACS Nano, 2018, 12 (2), 1508–1518
- Zahn, R., Osmanović, D., Ehret, S., Araya Callis, C., Frey, S., Stewart, M., You, C, Görlich, D, Hoogenboom, BW, Richter RP, eLife, 2016, 5, e14119
- A. Bestembayeva, A. Kramer, A. A. Labokha, D. Osmanović, I. Liashkovich, E. V. Orlova, I. J. Ford, G. Charras, A. Fassati and B. W. Hoogenboom, Nature Nanotechnology, 2015, 10, 60-65